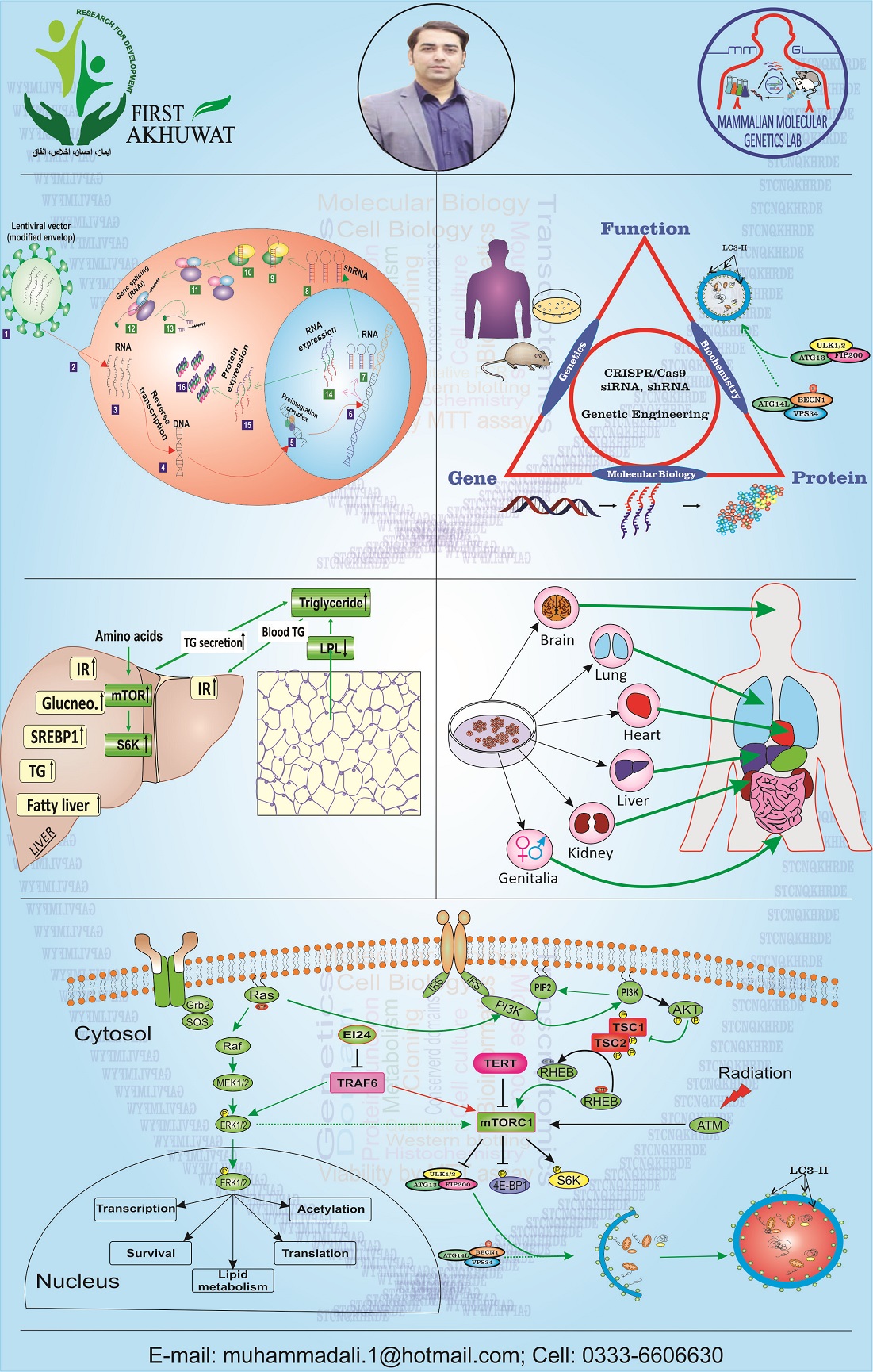
Introduction of MMGL:
We deals with the transcriptional, translational, and metabolic studies of diabetes, breast cancer, and liver injury. We utilized prokaryotic system, cell lines, mouse models, and human samples for our research work. During the diabetes and cancer that are due to uneven metabolism, we check the transcription of the genes and protein level. We observe different metabolic and cell survival pathways in liver and adipose tissues. Along with metabolic studies, study of signalling pathway is specialty of MMGL. At protein level, we are majorly interested in two pathways i.e. PI3K-AKT-mTORC1 and MAPK-ERK. The pathways are involved in diabetes and breast cancers respectively. For the study of these pathways, we use Western blotting. We use different tools of bioinformatics for designing of primers, analysis of data, and for phylogenetic analysis. We use NCBI, Ensemble, Primer quest, Mega 6.06, and SnapGene routinely in our lab.
Techniques used by MMGL
- Polymerase chain reaction; Site-directed mutagenesis, insertion, deletion (molecular cloning)
- Gene isolation and gene expression studies
- RNA extraction, cDNA synthesis, and qPCR
- Protein extraction, SDS-PAGE and Western blotting for cell signaling studies
- Design and making gene knockdown and knockout system using siRNA/shRNA and CRISPR/Cas9 respectively
- Invitro synthesis of guided RNA to use in CRISPR/Cas9 system
- The cell culture of all kinds of cell lines including primary, cancer, and embryonic stem cells
- A lentiviral system to knockdown/knockout genes from cell lines and mice for the functional study of proteins in-vivo
- Stable cell line generation by lentivirus-mediated gene knockdown
- MTT assay to check the viability of cells
- Colony formation assays to check the oncogenic potential of proteins in cancer cell lines.
- Mouse necropsy and organ collection for in-vivo studies
- RNA, DNA, and protein extraction and then expression test in different organs of mice
- Histochemistry analysis to check tissue morphology
- Generation of tissue and organ-specific KO and transgenic mice
- NCBI primer blast link for designing primers for genotyping and cloning
- DNA and protein homology using NCBI link, Muscle link, Cluster W link, T-coffee link
- Oncomine for the expression of genes in cancer tissues link
- Snap-Gene for cloning, genotyping, alignment, and map construction link
- Analysis of diversity among different members of a species using dendogram (Omega 6)
Research Interests of MMGL
- Bioinformatics analysis to find significance of genes involved in liver and breast cancers
- Insilico analysis to find binding of drugs with receptors
- Production of recombinant proteins (cell lines)
- Finding new pathways that control cell proliferation in liver and breast cancer (cell lines)
- Finding new signalling pathways/pathways involved in autophagy (cell lines)
- Finding novel pathways, including PI3K-AKT-mTORC1, that regulate type 2 diabetes (cell lines)
- Finding phytochemicals and drugs for the treatment of diabetes and liver injury (Cell lines)
- Finding phytochemicals and drugs for the treatment of diabetes and liver injury (mouse models)
- Signalling pathways involved in diabetes and liver injury (using mouse models)
- For Further details Visit: http://www.lifesciencesorg.com/